Cumulative SARS-CoV-2 Cases in the District of Columbia by Health Planning Neighborhoods
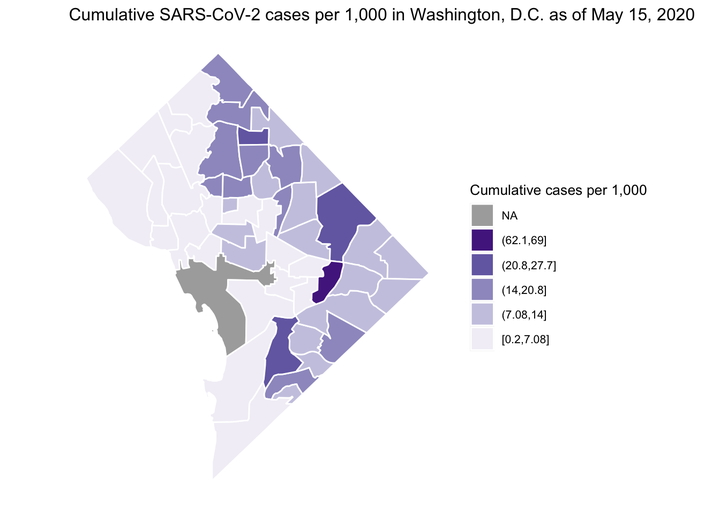 Image credit: Ian Buller
Image credit: Ian Buller
After moving to DC last year, PoPville has been a personal favorite for local scoop. A post on May 11, 2020 captured my attention. Molly Tolzmann zmotoly adjusted the daily coronavirus data publicly released by the DC Mayoral Office at the DC health planning neighborhood level by the 2018 American Community Survey (ACS) census tract data and demographic data from Open Data DC. This post is a replication of the data visualization using R and can be found on a public GitHub repository.
Imporant Notes:
- The following show cumulative case rate per 1,000 since the beginning of the COVID-19 outbreak in DC. Therefore, the the stats do not reflect the number of people currently infected with SARS-CoV-2.
- The following does not account for the degree of COVID-19 testing in each neighborhood.
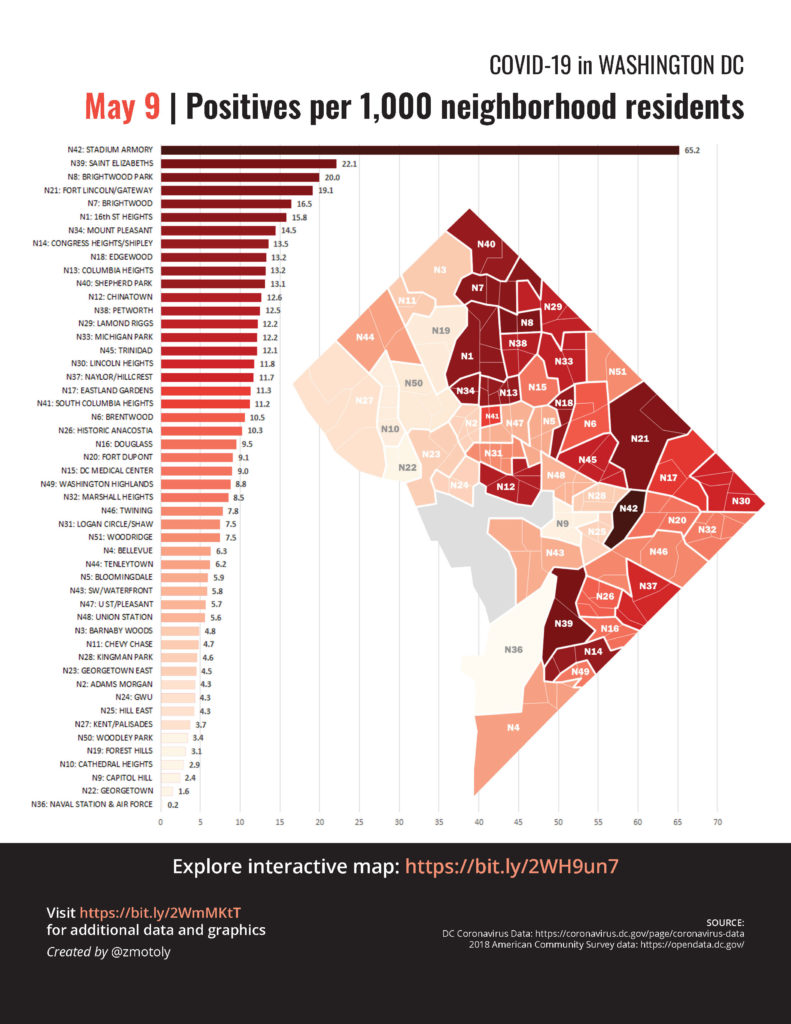
COVID-19 in DC by Molly Tolzmann
The following are the necessary packages and settings for the exercise.
# Packages
loadedPackages <- c("broom", "geojsonio", "ggplot2", "googlesheets4", "leaflet", "sp", "stringr", "widgetframe")
invisible(lapply(loadedPackages, require, character.only = T))
# Settings
googlesheets4::gs4_deauth() # no Google authorization necessary because we are not reading a public repoWe import the District of Columbia Health Planning Neighborhoods boundaries and Molly Tolzmann’s zmotoly collation of the cumulative cases from start of the SARS-CoV-2 outbreak. The latter is hosted on a public Google Sheet and is accessible in R using the googlesheets4 package. After cleaning up the column names of the disease data, we merge the two data sets together and spatially project the polygons contained in the data to EPSG:32618.
# District of Columbia Health Planning Neighborhoods
gis_path <- "https://opendata.arcgis.com/datasets/de63a68eb7674548ae0ac01867123f7e_13.geojson"
dc <- geojsonio::geojson_read(gis_path, what = "sp")
# District of Columbia SAR-CoV-2 Data prepared by @zmotoly
covid_path <- "https://docs.google.com/spreadsheets/d/1u-FlJe2B1rYV0obEosHBks9utkU30-C2TSkHka6AVS8/edit#gid=1923705378"
covid <- googlesheets4::read_sheet(ss = covid_path,
sheet = 2, # second sheet
skip = 2) # skip 1st row of annotation
names(covid) <- sub("\n", "", names(covid)) # remove extra line in column names
names(covid) <- gsub(" ", "_", names(covid)) # replace spaces with underscore
# Merge
dc_covid <- sp::merge(dc, covid, by.x = "CODE", by.y = "NB_Code")
# Spatial Projection
## UTM zone 18N (Washington, DC)
dc_covid_proj <- sp::spTransform(dc_covid, CRS("+init=EPSG:32618"))Static Map
We can then plot the cumulative rate using various plotting techniques. Here, I demonstrate the ggplot2 package. First, we convert the dc_covid_proj object of class SpatialPolygonsDataFrame to a class tibble.
# Uses ggplot2 package
## helpful material: https://cengel.github.io/rspatial/4_Mapping.nb.html
## Data preparation, ggplot2 requires a data.frame
dc_covid_df <- broom::tidy(dc_covid_proj) # convert to tidy data frame
dc_covid_proj$polyID <- sapply(slot(dc_covid_proj, "polygons"), function(x) slot(x, "ID")) # preserve polygon id
CoV_DC_df <- merge(dc_covid_df, dc_covid_proj, by.x = "id", by.y="polyID") # merge dataThen we can customize our plot in various ways. Here I present cumulative cases per 1,000 (May 15, since start of the outbreak) choosing a non-alarmist color palette from ColorBrewer2.0.
## Plot of cumulative cases per 1,000
ggplot2::ggplot() + # initialize ggplot object
ggplot2::geom_polygon( # make a polygon
data = CoV_DC_df, # data frame
ggplot2::aes(x = long, y = lat, group = group, # coordinates, and group them by polygons
fill = ggplot2::cut_interval(Cases_per_1000_May_15, 10)), # variable to use for filling
colour = "white") + # color of polygon borders
ggplot2::scale_fill_brewer("Cumulative cases per 1,000", # title of colorkey
palette = "Purples", # fill with brewer colors
na.value = "grey67", # color for NA (The National Mall)
direction = 1, # reverse colors in colorkey
guide = ggplot2::guide_legend(reverse = T)) + # reverse order of colokey
ggplot2::ggtitle("Cumulative SARS-CoV-2 cases per 1,000 in Washington, D.C. as of May 15, 2020") + # add title
ggplot2::theme(line = ggplot2::element_blank(), # remove axis lines
axis.text = ggplot2::element_blank(), # remove tickmarks
axis.title = ggplot2::element_blank(), # remove axis labels
panel.background = ggplot2::element_blank(), # remove background gridlines
text = ggplot2::element_text(size = 10)) + # set font size
ggplot2::coord_equal() # both axes the same scale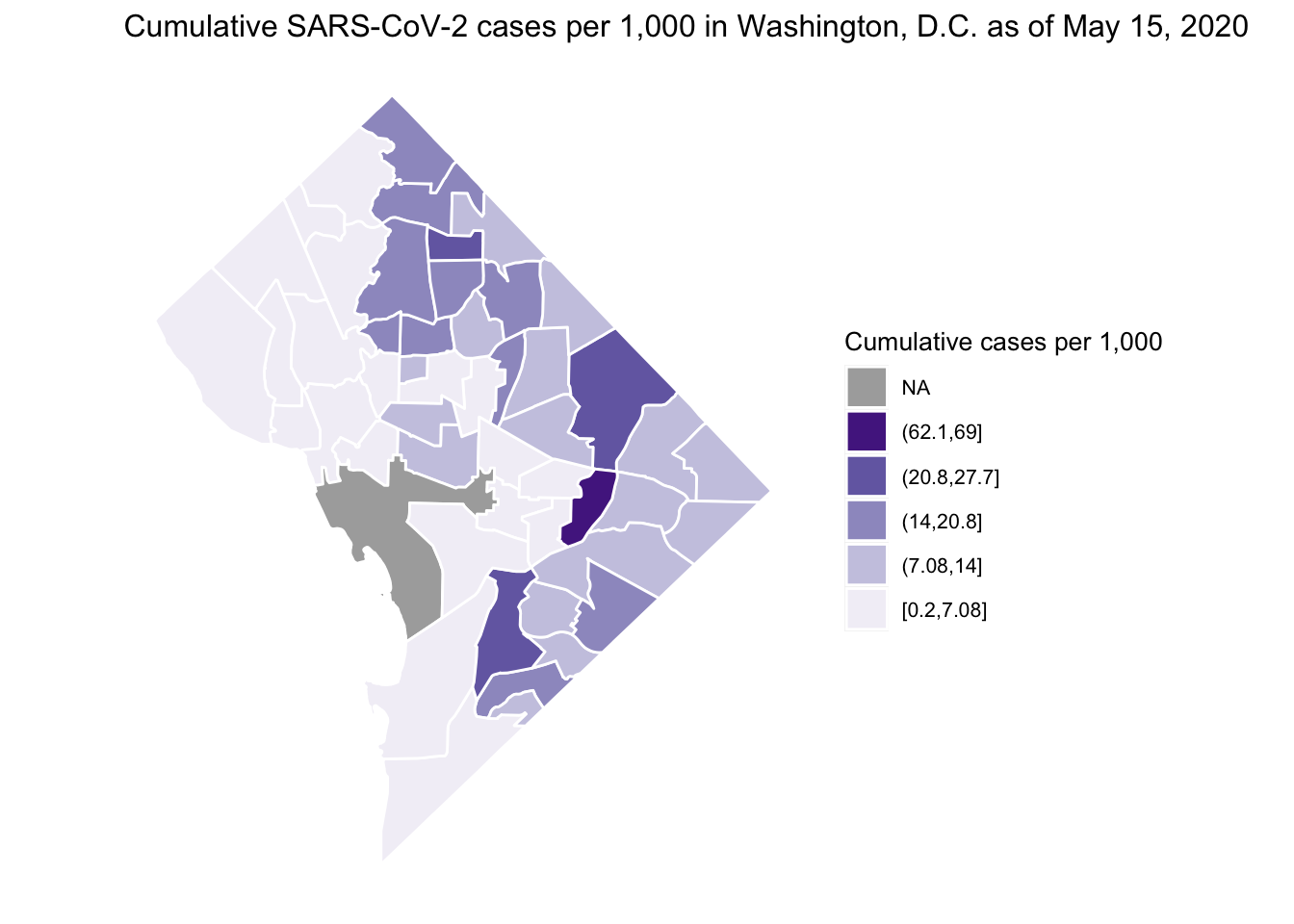
Interactive Map
In addition to a static map, the leaflet package provides capabilities to create an interactive map with customizable features such as basemaps and overlapping layers. First, we need to spatially project the data to EPSG:4326. We can also create custom popups when scrolling mouse over each neighborhood. Here, I use the same color palette as the static map and provide an example of the raw cumulative cases (May 15, 2020) in DC to demonstrate the layer overlapping feature of the leaflet package.
# Work with unprojected spatialpolygonsdataframe
## Project to WGS84 EPSG:4326
CoV_DC_WGS84 <- sp::spTransform(dc_covid_proj, CRS("+init=epsg:4326"))
## Create Popups
dc_health <- stringr::str_to_title(CoV_DC_WGS84$Neighborhood_Name)
dc_health[c(11,25,41,49)] <- c("DC Medical Center", "GWU", "National Mall", "SW/Waterfront" )
CoV_DC_WGS84$popup1 <- paste(dc_health, ": ",
format(round(CoV_DC_WGS84$Total_cases_May_15, digits = 0), big.mark = ",", trim = T),
" cumulative cases", sep = "")
CoV_DC_WGS84$popup2 <- paste(dc_health, ": ",
format(round(CoV_DC_WGS84$Cases_per_1000_May_15, digits = 0), big.mark = ",", trim = T),
" cumulative cases per 1,000", sep = "")
## Set Palettes
pal_cum <- leaflet::colorNumeric(palette = "Purples",
domain = CoV_DC_WGS84$Total_cases_May_15,
na.color = "#555555")
pal_rate <- leaflet::colorNumeric(palette = "Purples",
domain = CoV_DC_WGS84$Cases_per_1000_May_15,
na.color = "#555555")The following is code for the leaflet plot. I set the starting parameters including available basemaps and then add each layer of COVID-19 data as polygons. After specifying the legend for each layer I finish up with a mini map to show scale.
## Create leaflet plot
lf <- leaflet::leaflet(CoV_DC_WGS84) %>% # initial data
leaflet::setView(lng = -77, lat = 38.9, zoom = 11) %>% # starting coordinates
leaflet::addTiles() %>% # basemap
leaflet::addPolygons(data = CoV_DC_WGS84, color = "black", weight = 1, smoothFactor = 0.5, opacity = 1,
fillOpacity = 0.67, fillColor = ~pal_cum(Total_cases_May_15), popup = ~popup1,
highlightOptions = highlightOptions(color = "white", weight = 2, bringToFront = TRUE),
group = "Cumulative Cases") %>%
leaflet::addPolygons(data = CoV_DC_WGS84, color = "black", weight = 1, smoothFactor = 0.5, opacity = 1,
fillOpacity = 0.67, fillColor = ~pal_rate(Cases_per_1000_May_15), popup = ~popup2,
highlightOptions = highlightOptions(color = "white", weight = 2, bringToFront = TRUE),
group = "Cumulative Rate") %>%
leaflet::addLayersControl(overlayGroups = c("Cumulative Cases", "Cumulative Rate"), # layer selection
options = layersControlOptions(collapsed = FALSE)) %>%
addLegend("topright", pal = pal_cum, values = ~Total_cases_May_15, # legend for cases
title = "Cumulative Cases", opacity = 1, group = "Cumulative Cases") %>%
addLegend("topright", pal = pal_rate, values = ~Cases_per_1000_May_15, # legend for rate
title = "Cumulative Rate per 1,000", opacity = 1, group = "Cumulative Rate") %>%
leaflet::hideGroup(c("Cumulative Cases", "Cumulative Rate")) %>% # no data shown (default)
leaflet::addMiniMap(position = "bottomleft") # add mini map
widgetframe::frameWidget(lf, width='100%')As of May 15, 2020 the highest cumulative rate of SARS-CoV-2 cases have occurred in the Stadium Armory and Molly Tolzmann zmotoly noted the DC Jail is located in this neighborhood in her original post.
The provided maps are not intended to inform decision-making. Instead, I provide the the open-source code to download, manage, and visualize publicly available data. Future steps include linking other demographic information to each DC Health Planning Neighborhood (e.g., housing occupancy) and assessing their relationships with disease occurrence or creating an automatic workflow to update these figures daily.